Physical and chemical properties of DNA
DNA is a long polymer composed of repeating units, the nucleotides. A double-stranded DNA measured from 22 to 26 angstroms (2.2 to 2.6 nanometers) wide, and one unit (one nucleotide ) measures 3.3 Å (0.33 nm) long. Although each individual repeating unit is very small, DNA polymers can be molecules containing millions of huge nucleotides. For example, the longest human chromosome, the chromosome number 1, has approximately 220 million base pairs. In the organisms living DNA does not usually exist as a single molecule but as a pair of closely related molecules.
The two DNA strands are curled in on themselves forming a sort of spiral staircase, called a double helix. The model of double helix structure was proposed in 1953 by James Watson and Francis Crick (Article Molecular Structure of Nucleic Acids: A Structure for deoxyribose Nucleic Acid was released on April 25th of 1953 in Nature). The success of this model is its consistency with the physical and chemical properties of DNA. The study also showed that the complementarity of bases could be relevant in its replication, and also the importance of the base sequence as a carrier of genetic information. Each repeating unit, the nucleotide contains a segment of the support structure (sugar + phosphate), which holds the chain together, and a base, which interacts with the other DNA strand in the helix. In general, a base linked to a sugar is called a nucleoside and a base linked to a sugar and one or more phosphate groups is called a nucleotide. When many nucleotides are attached, as in DNA, the resulting polymer is called a polynucleotide.
Components
Support structure: The support structure of a strand of DNA consists of alternating units of groups phosphate and sugar. The sugar in DNA is a pentose, specifically, the deoxyribose.
Phosphoric acid:
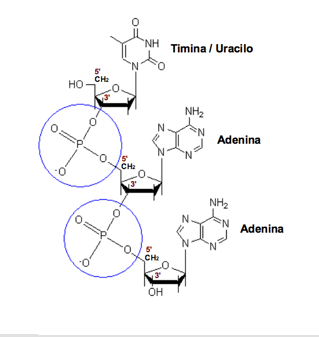
Phosphodiester bond. The phosphate group linking the carbon 5 'sugar of a nucleoside with carbon 3' of the next.
Its chemical formula is H 3 PO 4. Each nucleotide can contain one (monophosphate: AMP), two (diphosphate: ADP) or three (triphosphate: ATP) phosphoric acid groups, but as constituent monomers of nucleic acids only appear as nucleoside monophosphate.
Deoxyribose:
It is a monosaccharide of 5 atoms of carbon (a pentose) derived from the ribose, which forms part of the structure of DNA nucleotides. Its formula is C 5 H 10 O 4. One of the main differences between DNA and RNA is the sugar, as in the RNA of 2 - deoxyribose of DNA is replaced by a pentose Alternatively, the ribose.
The sugar molecules are joined together by phosphate groups that form phosphodiester bonds between carbon atoms third (3 ', "three prime") and fifth (5', "five prime") of two adjacent sugar rings . The formation of links asymmetric means that each strand of DNA has a direction. In a double helix, the direction of the nucleotides on one strand (3 '→ 5') is opposite to the direction in the other strand (5 '→ 3'). This organization of the DNA strands is called antiparallel, are chains parallel but opposite directions. Similarly, the asymmetric ends of DNA strands are called the 5 '( "five prime") and the 3' ( "three prime") respectively.
Nitrogenous bases:
The four nitrogenous bases that are majority in DNA are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). Each of these four bases is attached to the structure of sugar-phosphate through the sugar to form the complete nucleotide (base-sugar-phosphate). The bases are heterocyclic compounds and aromatic with two or more atoms of nitrogen, and within the majority bases are classified into two groups: the purine bases or purines (adenine and guanine), derived from the purine and formed by two rings together each other, and the pyrimidine bases and pyrimidine (cytosine and thymine), derived from the pyrimidine ring with a single. In the nucleic acids A fifth pyrimidine base, called uracil (U), which normally takes the place of thymine in RNA and differs from it in lacking a methyl group in the ring. Uracil is not usually found in DNA, appears only rarely as a waste product of the degradation of cytosine deamination by oxidative processes.

Thymine: 2, 4-dioxo, 5-methylpyrimidine.
Thymine:
In the genetic code is represented by the letter T. It is a pyrimidine derivative with an oxo group in positions 2 and 4, and a methyl group at position 5. Form the nucleoside thymidine (deoxythymidine always as shown only in DNA) and the nucleotide thymidylate and thymidine monophosphate (dTMP). In DNA, thymine always pairs with adenine of the complementary strand by 2 hydrogen bonds, T = A. Its chemical formula is C 5 H 6 N 2 O 2 and nomenclature 2, 4-dioxo, 5-methylpyrimidine.

Cytosine: 2-oxo-4-aminopyrimidine.
Cytosine:
In the genetic code is represented by the letter C. It is a pyrimidine derivative with an amino group in position 4 and an oxo group in position 2. Form the nucleoside cytidine (deoxycytidine in DNA) and the nucleotide citidilato or (deoxy) cytidine monophosphate (dCMP in DNA into RNA CMP). Cytosine always pairs in DNA with guanine of the complementary strand by a triple bond, C ≡ G. Its chemical formula is C 4 H 5 N 3 O and nomenclature 2-oxo, 4 aminopyrimidine. Its molecular weight is 111.10 atomic mass units. Cytosine was discovered in 1894 when tissue was isolated in thymus of ram.

Adenine: 6-aminopurine.
Adenine:
In the genetic code is represented by the letter A. It is a purine derivative of an amino group in position 6. Form the nucleoside adenosine (deoxyadenosine in DNA) and the nucleotide adenylate or (deoxy) adenosine monophosphate (dAMP, AMP). In DNA always pairs with thymine on the complementary strand by 2 hydrogen bonds, A = T. Its chemical formula is C 5 H 5 N 5 and 6-aminopurine nomenclature. Adenine, thymine with, was discovered in 1885 by German physician Albrecht Kossel.
Guanine: 6-oxo, 2-aminopurine.
Guanine:
In the genetic code is represented by the letter G. It is a purine derivative with an oxo group in position 6 and an amino group in position 2. Form the nucleoside (deoxy) guanosine and the nucleotide guanylate or (deoxy) guanosine monophosphate (dGMP, GMP). The guanine always pairs in DNA with the complementary strand cytosine through three hydrogen bonds, G ≡ C. Its chemical formula is C 5 H 5 N 5 O and nomenclature 6-oxo, 2-aminopurine.
There are also other nitrogen bases (nucleobases calls minority), derived from natural or synthetic form another majority basis. What are eg hypoxanthine, relatively abundant in the tRNA, or caffeine, both derived from adenine, others, such as acyclovir, derived from guanine, are synthetic analogues used in antiviral therapy, sometimes as one of the derivatives of uracil-antitumour.
The nitrogenous bases are a number of characteristics that give them specific properties. An important feature is its aromatic character, due to the presence in the ring double bonds in conjugated position. This gives them the ability to absorb light in the ultraviolet the spectrum around 260 nm, which can be exploited to determine the extinction coefficient of DNA and find the existing concentration of nucleic acids. Another feature is that they present tautomerism or isomerism of functional groups due to an atom of hydrogen bound to another atom can migrate to a neighboring position, in the nitrogenous bases are two types of tautomerism: lactim-lactam tautomerism, where hydrogen migrates from nitrogen to oxygen of the oxo group (lactam form) and vice versa (as lactim) and primary amine-imine tautomerism, where hydrogen can be making the amine group (primary amine form) or migrate to adjacent nitrogen (imine form). Adenine can only present amine imine tautomerism, thymine and uracil are double lactam tautomerism-lactim, and guanine and cytosine may have both. Furthermore, and even if the molecules nonpolar and nitrogen bases have sufficient polar character to establish hydrogen as atoms are very electronegative (nitrogen and oxygen) presenting partial negative charge, and hydrogen atoms with partial positive charge so that form dipoles which allow to form these weak bonds.
It is estimated that the human genome haploid has some 3,000 million base pairs. To indicate the size of DNA molecules indicates the number of base pairs, and as deriving from two drives are widely used measure, the kilobase (kb), which is 1,000 base pairs, and megabase (Mb) equivalent to one million base pairs.
Base pairing

A ≡ C base pair G with three hydrogen bonds.

A = T A pair with two hydrogen bonds. Hydrogen bonds are shown as dashed lines.
The DNA double helix is stable through the formation of hydrogen bonds between the bases associated with each of the two strands. For the formation of a hydrogen bond of the bases must submit a "donor" of hydrogen with a hydrogen atom with partial positive charge (-NH 2 or-NH) and the other a group basis must submit "acceptor" of hydrogen with a charged atom electronegativity (C = O or N). Hydrogen bonds are bonds weaker than typical chemical bonds-covalent, such as connecting the atoms in each strand of DNA, but stronger than individual hydrophobic interactions, Van der Waals bonds, and so on. As hydrogen bonds are not covalent bonds can break and form again relatively easily. For this reason, the two strands of the double helix can be separated like a zipper, either by mechanical force or high temperature. The double helix is stabilized also by the effect hydrophobic and stacking, which are not influenced by the sequence DNA base.
Each type of base on one strand forms a bond with only a base rate in the other strand, which is called "complementarity of the bases. Accordingly, form bonds with purines pyrimidines, so that A binds only to T and C only with G. The organization of two unpaired nucleotides along the double helix is called base pairing. This pairing corresponds to the observation already made by Erwin Chargaff (1905-2002), [29] which showed that the amount of adenine was very similar to the amount of thymine, and that the amount of cytosine was equal to the amount of guanine in DNA. As a result of this complementarity, all information contained in the double-stranded sequence of the DNA helix is duplicated on each strand, which is essential during the process of DNA replication. Indeed, this reversible and specific interaction between complementary base pairs is critical for all functions of DNA in living organisms.
As mentioned previously, the two types of base pairs form different numbers of hydrogen bonds: A = T form two hydrogen bonds and C ≡ G form three hydrogen bonds . The GC base pair is therefore stronger than the AT base pair. As a result, both the percentage of GC base pairs as the total length of the double helix of DNA determine the strength of association between the two strands of DNA. The long double helix of DNA with high GC content have more strongly interacting strands that short double helices with high AT content. [30] For this reason, areas of the double helix of DNA that need to be separated easily tend to have high in AT, such as the sequence TATAAT Pribnow box in some promoters. In the laboratory, the strength of this interaction can be measured for the temperature required to break hydrogen bonds, the melting temperature (aka T m value, the English melting temperature). When all the base pairs in a double helix melt, the strands are separated into two strands in solution completely independent. These molecules single-stranded DNA does not have a single common form, but some conformations are more stable than others.
Other types of base pairs

Base pair A = T Watson-Crick type. In blue the hydrogen donor and acceptor in red.

Base pair of type A = T Watson-Crick reverse. In blue the hydrogen donor and acceptor in red. Note that the pyrimidine has suffered a 180 º about the axis of the carbon 6.
There are different types of base pairs that can be formed according to how they form hydrogen bonds. Those who are in the double helix of DNA are called pairs of Watson-Crick bases, but there are other possible pairs, as called Hoogsteen and Wobble or oscillating, which can occur in particular circumstances. Moreover, for each type there to turn back the same pair, ie, that is if the pyrimidine base is rotated 180 degrees on its axis.
Watson-Crick (base pairs of the double helix): the purine base groups involved in hydrogen bonding are those that correspond to positions 1 and 6 (N-NH 2 acceptor and donor if the purine is an A ) and the pyrimidine base groups which are at positions 3 and 4 (-NH donor and C = O acceptor if the pyrimidine is a T). Base pair in Watson-Crick groups participate reverse positions 2 and 3 of the pyrimidine base .
Hoogsteen: in this case changing the purine base groups, which offers a different face (positions 6 and 7) and form links with groups of pyrimidines from positions 3 and 4 (as in Watson-Crick). There may also be reverse Hoogsteen. With this type of link can bind A = U (Hoogsteen and reverse Hoogsteen) and A = C (reverse Hoogsteen).
Wobble or oscillating: This type of link allows you to join guanine and cytosine with a double bond (G = T). The purine base (G) is liaising with groups in positions 1 and 6 (as in Watson-Crick) and pyrimidine (T) with groups 2 and 3 positions. This type of link would not work with A = C, because the 2 would be facing the 2 acceptors and donors, and can only be given in the reverse case. We found pairs of oscillating type RNA during mating of codon and anticodon. With this type of link can bind G = U (swing and reverse swing) and A = C (reverse swing).
In total, as tautomeric majority, there are 28 possible pairs of nitrogenous bases: 10 possible base pair purine-pyrimidine (2 Watson-Crick pairs and 2 reverse Watson Crick, 1 pair Hoogsteen and reverse Hoogsteen 2 pairs, 1 pair oscillating 2 pairs oscillating back), 7 pairs homo purine-purine (A = A, G = G), 4 pairs A = G and 7 pyrimidine-pyrimidine pairs.